BACKGROUND
BACKGROUND
Many species of fungi capable of growing in grain and animal feed produce secondary metabolites called mycotoxins.
Mycotoxins, such as fumonisin FB1 (FB1) and T-2 toxin (Figure 1), are the biggest challenge for animal feed producers and therefore, regular monitoring and remediation is necessary, as these toxins can be very detrimental to humans and animals.
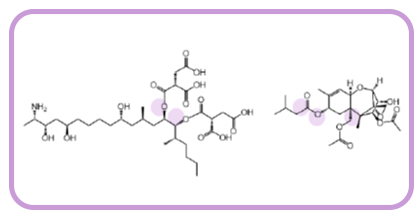
Figure 1. FB1 and T-2 toxin. The sites that can be potentially attacked by enzymes are highlighted in lilac.
 The economic loss attributed to mycotoxins in the agri-food sector is estimated to be billions of dollars, so the economic potential of protection or mycotoxin removal measures is correspondingly high.
The economic loss attributed to mycotoxins in the agri-food sector is estimated to be billions of dollars, so the economic potential of protection or mycotoxin removal measures is correspondingly high.
There are already different measures available today, such as mechanical, thermic, or chemical approaches.
 However, these approaches have, as an associated disadvantage, the possibility of generating unknown or unintended side products that may turn out to be as dangerous as the original toxin.
However, these approaches have, as an associated disadvantage, the possibility of generating unknown or unintended side products that may turn out to be as dangerous as the original toxin.
Enzymes are proteins that catalyze biochemical reactions that have been making life possible since 3.8 billion years ago.
 Scientists have been able to “domesticate” them in such a way that enzymes are now key players for the circular economy and for the processing, manufacturing, and consuming everyday products, both for humans and animals2.
Scientists have been able to “domesticate” them in such a way that enzymes are now key players for the circular economy and for the processing, manufacturing, and consuming everyday products, both for humans and animals2.
OBJECTIVE
OBJECTIVE
Repowering the agri-food industry with enzymes capable of efficiently removing mycotoxins can contribute to keeping our food and feed safe and reducing carryover of mycotoxins from animals to humans.
However, the option to find such new enzymes, particularly those to degrade mycotoxins such as FB1 and T-2 toxin is challenging, costly, and time-consuming.
 Indeed, only a few enzymes are known that degrade both toxins1.
Indeed, only a few enzymes are known that degrade both toxins1.
 In this contribution, we provide a comprehensive overview of our synergetic platform to better bio-prospect environmental metagenomes for such novel rare enzymes.
In this contribution, we provide a comprehensive overview of our synergetic platform to better bio-prospect environmental metagenomes for such novel rare enzymes.
We further provide experimental evidence that demonstrates the efficient degradation of FB1 and T-2 toxin by two novel hydrolases retrieved through naïve and bioinformatic screens.
MATERIALS & METHODS
MATERIALS & METHODS
The option to find new enzymes, although challenging, costly (€30k/enzyme), and time-consuming (15 months/enzyme), it is realistic given the recent technological advances.
To fully exploit the potential of sequencing data and as a source of new enzymes it is essential to combine:
- ⇰ Bioinformatics
- ⇰ Machine-learning
- ⇰ Accurate protein structure prediction
- ⇰ Data-driven artificial intelligence techniques
- ⇰ Naïve screens with appropriate substrates
In order to access new enzymes capable of degrading mycotoxins, we have established a comprehensive and synergetic platform to better bio-prospect environmental metagenomes for novel rare enzymes (Figure 2).
It consists in the following steps:
- 1. Extraction and sequencing of DNA from microbial communities (the so-called metagenome) of a large number of environmental samples.
- 2. Bioinformatic processing of the sequences representing the metagenomes to filter for those enzymes in the metagenome that have the potential to degrade FB1 and T-2 toxin, namely, hydrolases.
- 3. Computational docking analysis to further provided computational quantum/molecular mechanics (QM/MM) analysis to identify, by quantifying the catalytic events, which of the selected enzymes can bind and potentially degrade FB1 and T-2 toxin.
- 4. Finally, the experimental validation: gene synthesis, production, purification, and characterization.
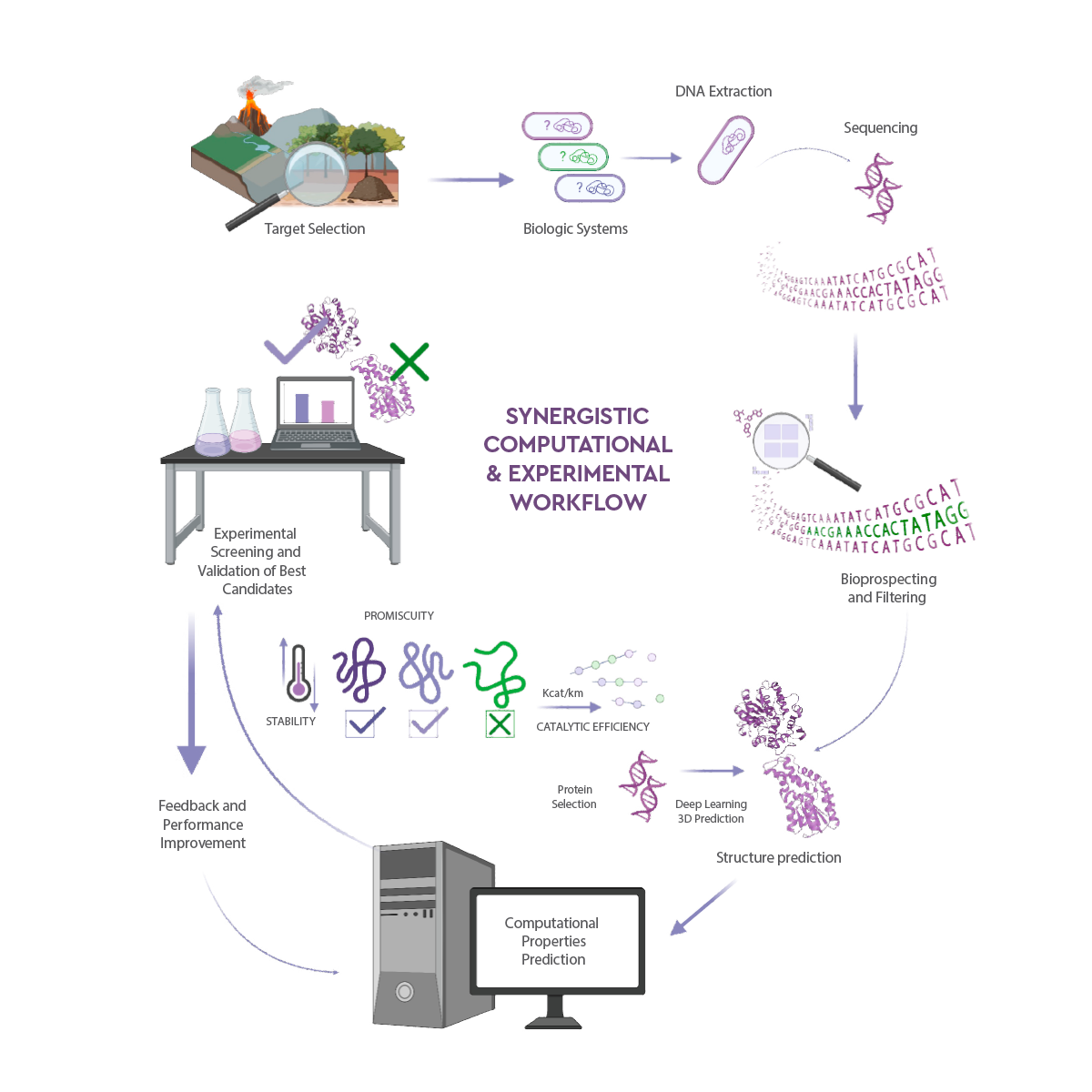
Figure 2. Schematic workflow for the bioprospecting of enzymes capable of degrading toxins. Shown are the steps related to extraction, sequencing, assembling, annotation, and virtual screening of new enzymes from the metagenomes of environmental samples, followed by their accurate protein structure prediction, high throughput characterization, and iterative improvement by engineering (created with BioRender.com).
RESULTS
RESULTS
Through the application of our synergetic platform, we have successfully bio-prospected environmental metagenomes and found several FB1 and T-2 toxin degrading hydrolases, to highlight:
- ✓ A family VIII esterase, isolated from the metagenomic DNA of microbial communities inhabiting the seashore area of Milazzo Harbor (Sicily, Italy), capable of degrading T-2 toxin (Figure 3) without producing intermediate HT-2 toxin1.
- ✓ An esterase capable of degrading 2-10 ppm FB1 (100% degradation) after a 5-minute reaction time at concentrations as low as 5 μg/ml at 37-90 °C3.
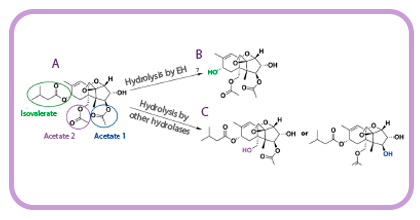
Figure 3. (A) Structure of T-2 toxin. Different ester bonds that can be hydrolyzed are colored. Product(s) obtained by the degradation of T-2 toxin through the isovalerate group by the esterase EH7 (B) and by the hydrolysis of both acetates by different hydrolases (C).
Authors
Manuel Ferrer1, David Almendral1, Jog Raj2, Hunor Farkaš2, and Marko Vasiljević2
1Department of Applied Biocatalysis, ICP, CSIC, Marie Curie 2, Madrid, Spain
2PATENT CO, DOO., Vlade Ćetković 1A, 24 211 Mišićevo, Serbia
1. Cea-Rama I, Coscolín C, Gonzalez-Alfonso JL, Raj J, Vasiljević M, Plou FJ, Ferrer M, Sanz-Aparicio J. FEBS J. 2022;289(21):6714-6730.
2. Molina-Espeja, P. et al. Oxford Open Climate Change, 2023; kgad003, https://doi.org/10.1093/oxfclm/kgad003
3. Fumonisin removal by enzymatic detoxification – EP1641.1745




 Micotoxicosis prevention
Micotoxicosis prevention