Al-Zahraa Mamdouh1† and Eman Zahran2*†
1Fish Diseases Department, National Institute of Oceanography and Fisheries (NIOF), Egypt
2Department of Aquatic Animal Medicine, Faculty of Veterinary Medicine, Mansoura University, Mansoura 35516, Egypt
†These authors share first authorship
*Corresponding author: [email protected] ORCID: 0000-0003-2212-3688
Fungi in food or feed produce mycotoxins that can harm humans and animals.

The hepatotoxic, nephrotoxic, carcinogenic, and mutagenic effects of mycotoxins, as well as their impact on the immune and reproductive systems, make them a major concern when it comes to food and feed safety.
Traditional physical and chemical mycotoxin detoxification methods for food and feed can result in the loss of nutrients, the presence of reagent residues, and pollution of the environment. Thus, mycotoxin contamination control requires new detoxification methods.
In this scenario, microbial detoxification technology has become increasingly popular for degrading mycotoxins in food and feed due to its high efficiency, low toxicity, and strong specificity without damaging nutrients.
The scourge of mycotoxins in food and feed
Mycotoxins are toxic chemical compounds produced as secondary metabolites by filamentous fungi that contaminate food/ feedstuffs or the raw materials used to make them (Tola and Kebede, 2016).
Aspergillus, Penicillium y Fusarium represent the main genera of mycotoxigenic fungi, in addition to Trichoderma, Trichothecium and Alternaria (Ashiq, 2015).
The growth of moulds and the production of mycotoxins is determined by certain environmental conditions, including:
 Humidity levels (>70%)
Humidity levels (>70%) Temperature (20–30 °C)
Temperature (20–30 °C) Moisture content of the plants or crops (> 15%)
Moisture content of the plants or crops (> 15%) Mechanical damage and damage to crops by insects and the use of pesticides
Mechanical damage and damage to crops by insects and the use of pesticides Plant diversity
Plant diversity Fungal spore load
Fungal spore load
(Streit et al., 2012)

 The proliferation of various fungi in agricultural products reduces yield and quality with significant economic losses (Adejumo and Adejoro, 2014).
The proliferation of various fungi in agricultural products reduces yield and quality with significant economic losses (Adejumo and Adejoro, 2014).
Approximately 25% of the world’s crops are contaminated with mycotoxins yearly (Alshannaq and Yu, 2017).

In addition, the annual losses caused by the contamination of feedstock with mycotoxins in the husbandry (Ogunade et al., 2018) and aquaculture (Anater et al., 2016) industries reaches 1 billion metric tons of food (Smith et al., 2016).
Annually, the United States faces an estimated $932 million in economic losses due to af aflatoxins, fumonisins, and deoxynivalenol (Bhatnagar et al., 2004).
The contamination of food and feedstuff with mycotoxins diminishes their nutritional value and represents a potential risk for human, animal, and fish health (Koletsi et al., 2021).
Bioremediation – A promising strategy against mycotoxins
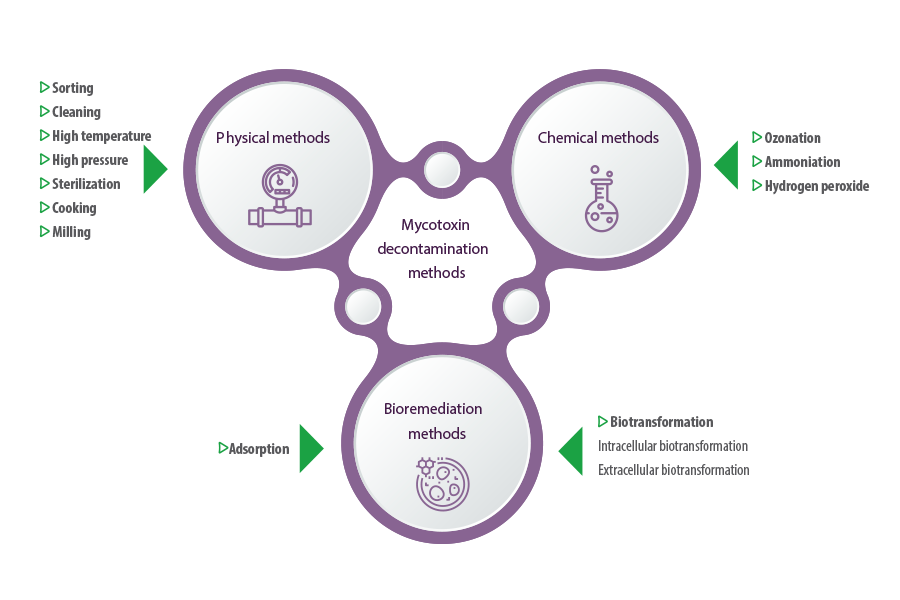
Methods of mycotoxin decontamination in feedstuff have been limited to physical and chemical techniques for a long time.
- ⇰ Physical methods include sorting, cleaning, high temperature, high pressure, and other practices, such as sterilization, cooking, and milling (Grenier et al., 2014; Pankaj et al., 2018).
- ⇰ Chemical methods include the use of chemical agents to reduce or convert mycotoxins into less toxic byproducts through ozonation, ammoniation and the use of hydrogen peroxide (Čolović et al., 2019).
While these methods have effectively reduced mycotoxin contamination, they have limitations, including not being eco-friendly, high operational costs, and less reliable results. Additionally, they may decrease the quality or nutritional value of the food (Čolović et al., 2019; Pankaj et al., 2018).
 Bioremediation is a promising emerging technology for mycotoxin degradation and detoxification based on the use of biological entities, such as bacteria or fungi, to detoxify or remove environmental toxins (Chandra, 2015).
Bioremediation is a promising emerging technology for mycotoxin degradation and detoxification based on the use of biological entities, such as bacteria or fungi, to detoxify or remove environmental toxins (Chandra, 2015).Converting mycotoxins into non-toxic compounds using microbial agents could be the best alternative to classic decontamination methods.
 Bioremediation is considered one of the most promising approaches because it has high degradation efficiency and causes fewer hazards in the environment and less reduction in the quality of food and feed products.
Bioremediation is considered one of the most promising approaches because it has high degradation efficiency and causes fewer hazards in the environment and less reduction in the quality of food and feed products.
Food decontamination by microbial enzymes is even more favorable as there has been significant development of microbial and genetic engineering technologies and rapid progress in the study of mycotoxin degradation by key enzymes and heterologously expressed degrading enzymes.
In addition, these enzymes are substrate-specific, effective, and environmentally friendly, and can generally maintain food value without causing loss of nutrients and food texture (Wang y Xie, 2020).
Mechanisms of mycotoxin bioremediation
Microbes can degrade mycotoxins either through adsorption or biotransformation.
- ⇰ Adsorption involves the physical binding of mycotoxins to polysaccharides and proteins on the outer cell structures of the microbe, forming a toxin-microbe complex that can be excreted (Nazareth et al., 2019; Sadiq et al., 2019).
- ⇰ Biotransformation involves the conversion of mycotoxins into non-toxic or less toxic compounds using microbes or their intracellular and extracellular enzymes (Alberts et al., 2009; Vega et al., 2017).
Intracellular biodegradation is based on the normal metabolic treatment of molecules by microbes inside the cell (Sandlin et al., 2022), whereas extracellular biotransformation occurs through the production of stable enzymes that can degrade mycotoxins.
This basic technology produces several commercial products (Moll, 2019).
Common mycotoxins and their bioremediation
AFLATOXINS
Aflatoxins (AFB) are the most hazardous, toxic, and studied mycotoxins.
Produced mainly by Aspergillus flavus and A. parasiticus and, to a lesser extent, by A. nomius, A. bombycis, A. pseudotamari, and A. ochraceoroseus (Varga et al., 2011), they are classified according to their blue or green fluorescence under ultraviolet light into AFB1, AFB2, AFG1 y AFG2 (Dhanasekaran et al., 2011).
They are produced in various feedstuffs, including maize, ground nuts, rice and sorghum cottonseed, spices, cereals, soybean, cocoa, and meat (Patriarca y Pinto, 2017; Vila-Donat et al., 2018).
Aflatoxins have hepatotoxic, teratogenic, mutagenic (Kucukcakan y Hayrulai-Musliu, 2015; Rieswijk et al., 2016; Rotimi et al., 2017) and immunosuppressive properties (Gao et al., 2023; Gong et al., 2016; Mohsenzadeh et al., 2016).
 AFB1 is one of the most potent hepatocarcinogens and a major contributor to the worldwide occurrence of hepatocarcinoma (HCC) (Kew, 2013).
AFB1 is one of the most potent hepatocarcinogens and a major contributor to the worldwide occurrence of hepatocarcinoma (HCC) (Kew, 2013).Bioremediation of aflatoxins by microbial adsorption
There are many studies on aflatoxin biodegradation, either by microbial agents or their enzymes.
Among the microorganisms that can bind and remove aflatoxins through adsorption, probiotics such as lactic acid bacteria and yeasts are studied the most because they can be added during food fermentation to degrade aflatoxins in the production process (Shetty y Jespersen, 2006).
LACTIC ACID BACTERIA
 Lactobacillus rhamnosus (GG) removed 54% of AFB1 from chicken intestinal juice medium in 1 min (El-Nezami et al., 2000).
Lactobacillus rhamnosus (GG) removed 54% of AFB1 from chicken intestinal juice medium in 1 min (El-Nezami et al., 2000). Lactic acid bacteria reduced, in vitro, AFB1 transport, metabolism, and toxicity in Caco-2 cells (Gratz et al., 2007).
Lactic acid bacteria reduced, in vitro, AFB1 transport, metabolism, and toxicity in Caco-2 cells (Gratz et al., 2007). The binding of 20 strains of lactic acid bacteria and bifidobacteria to AFB1 in a contaminated solution was studied. Two Lactobacillus amylovorus strains and one L. rhamnosus strain bound more than 50%of AFB1 at 72 h (Peltonen et al., 2001).
The binding of 20 strains of lactic acid bacteria and bifidobacteria to AFB1 in a contaminated solution was studied. Two Lactobacillus amylovorus strains and one L. rhamnosus strain bound more than 50%of AFB1 at 72 h (Peltonen et al., 2001). Lactobacillus plantarum C88 isolated from Chinese traditional fermented foods reduced the toxicity of AFB1 in mice, demonstrating that the mechanism behind detoxification was a strong binding capacity that resulted in an increased amount of unabsorbed AFB1 in the faeces (Huang et al., 2017).
Lactobacillus plantarum C88 isolated from Chinese traditional fermented foods reduced the toxicity of AFB1 in mice, demonstrating that the mechanism behind detoxification was a strong binding capacity that resulted in an increased amount of unabsorbed AFB1 in the faeces (Huang et al., 2017).
 L. plantarum MON03 (LP) isolated from Tunisian artisanal butter was found to display a significant binding ability to AFB1 and AFM1 in phosphate buffer saline (PBS) by 82% and 89%, respectively, within 24 h of incubation with the ability to tolerate gastric acidity and strong hydrophilic cells surface properties, and adherence efficacy to Caco-3 cells in vitro (Jebali et al., 2015).
L. plantarum MON03 (LP) isolated from Tunisian artisanal butter was found to display a significant binding ability to AFB1 and AFM1 in phosphate buffer saline (PBS) by 82% and 89%, respectively, within 24 h of incubation with the ability to tolerate gastric acidity and strong hydrophilic cells surface properties, and adherence efficacy to Caco-3 cells in vitro (Jebali et al., 2015). L. plantarum MON03 and L. rhamnosus GAF01 were isolated and tested for their abilities to eliminate AFM1 from PBS and reconstituted milk and it was found that the two strains removed AFM1, both in PBS and skimmed milk after 0, 6, and 24 h at 37 °C, and their binding abilities ranged from 16.1–78.6% and 15.3–95.1%, respectively (Abbès et al., 2013).
L. plantarum MON03 and L. rhamnosus GAF01 were isolated and tested for their abilities to eliminate AFM1 from PBS and reconstituted milk and it was found that the two strains removed AFM1, both in PBS and skimmed milk after 0, 6, and 24 h at 37 °C, and their binding abilities ranged from 16.1–78.6% and 15.3–95.1%, respectively (Abbès et al., 2013).
YEASTS
 Saccharomyces cerevisiae bound 90.3 ± 0.3% and 92.7 ± 0.7% of AFM1 for 30 and 60 min, respectively, in milk. In comparison, it was able to bind 100% AFM1 within 60 min when combined with a pool of other lactic acid bacteria strains (L. rhamnosus, Lactobacillus delbrueckii spp. bulgaricus and Bifidobacterium lactis) (Corassin et al., 2013).
Saccharomyces cerevisiae bound 90.3 ± 0.3% and 92.7 ± 0.7% of AFM1 for 30 and 60 min, respectively, in milk. In comparison, it was able to bind 100% AFM1 within 60 min when combined with a pool of other lactic acid bacteria strains (L. rhamnosus, Lactobacillus delbrueckii spp. bulgaricus and Bifidobacterium lactis) (Corassin et al., 2013). S. cerevisiae bound 60% and 40% AFB1 in beer fermentation residue at pH 3 and 6.5, respectively (Campagnollo et al., 2015).
S. cerevisiae bound 60% and 40% AFB1 in beer fermentation residue at pH 3 and 6.5, respectively (Campagnollo et al., 2015). The ability of S. cerevisiae–based products from sugar cane fermentation (autolyzed yeast and dried yeast) to bind AFB1 in PBS was evaluated and it was reported that autolyzed yeast bound 90.4% -97.5% AFB1. The dried yeast binding capacity was 96.5-99.3% (Gonçalves et al., 2015).
The ability of S. cerevisiae–based products from sugar cane fermentation (autolyzed yeast and dried yeast) to bind AFB1 in PBS was evaluated and it was reported that autolyzed yeast bound 90.4% -97.5% AFB1. The dried yeast binding capacity was 96.5-99.3% (Gonçalves et al., 2015).
 Chlebicz and Śliżewska (2020) studied the ability of 12 strains of Lactobacillus sp. and six strains of S. cerevisiae yeast to bind AFB1, DON, fumonisin B1, and T-2 toxin.
Chlebicz and Śliżewska (2020) studied the ability of 12 strains of Lactobacillus sp. and six strains of S. cerevisiae yeast to bind AFB1, DON, fumonisin B1, and T-2 toxin.-
They found that all strains detoxified the mycotoxins, with the highest reduction in concentration for the FB1 and FB2 mixture, ranging between 62 and 77% for bacterial strains and 67–74% for yeast, and DON being the most resistant mycotoxin. Its concentration was reduced by 19–39% by Lactobacillus sp strains and 22–43% by yeast after 24 h of incubation while high detoxification rates for AFB1 and T-2 toxin were 60% and 61% by Lactobacillus, respectively, and 65% and 69% by yeast, respectively.
 S. cerevisiae NRRL Y-1089 had the ability to remove 87.20% AFB1 after 30 minutes of incubation and the binding improved by increasing the incubation time (Hathout y Abdel-Nasser, 2022).
S. cerevisiae NRRL Y-1089 had the ability to remove 87.20% AFB1 after 30 minutes of incubation and the binding improved by increasing the incubation time (Hathout y Abdel-Nasser, 2022).
Bioremediation of aflatoxins by microbial degradation
BACTERIA
 Myxococcus fulvus ANSM068 isolated from deer faeces transformed 80.7% AFB1 in liquid VY/2 medium after incubation at 30 °C for 72 h (Guan et al., 2010).
Myxococcus fulvus ANSM068 isolated from deer faeces transformed 80.7% AFB1 in liquid VY/2 medium after incubation at 30 °C for 72 h (Guan et al., 2010). Bacillus velezensis DY3108 degraded AFB1 by 91.5% with an optimal temperature of 80°C and a broad pH (4.0 to 11.0) with the highest degradation (94.70%) at pH 8.0 (Shu et al., 2018).
Bacillus velezensis DY3108 degraded AFB1 by 91.5% with an optimal temperature of 80°C and a broad pH (4.0 to 11.0) with the highest degradation (94.70%) at pH 8.0 (Shu et al., 2018). Bacillus licheniformis CFR1 degraded AFB1 by 94.7% after 24 h of incubation with an optimal temperature of 37 °C and pH 7 (Rao et al., 2017).
Bacillus licheniformis CFR1 degraded AFB1 by 94.7% after 24 h of incubation with an optimal temperature of 37 °C and pH 7 (Rao et al., 2017).
 Stenotrophomonas acidoaminiphila CW117 degraded 76.7% of the AFB1 under high temperatures (reached 90 ⁰C) and at pH 6-7, and the degradation was dose and time-dependent (Cai et al., 2020).
Stenotrophomonas acidoaminiphila CW117 degraded 76.7% of the AFB1 under high temperatures (reached 90 ⁰C) and at pH 6-7, and the degradation was dose and time-dependent (Cai et al., 2020). A non-pathogenic Escherichia coli CG1061 eliminated AFB1 with a 93.7% degradation rate at an optimal temperature of 55 °C and pH 8.5 (Wang et al., 2019a).
A non-pathogenic Escherichia coli CG1061 eliminated AFB1 with a 93.7% degradation rate at an optimal temperature of 55 °C and pH 8.5 (Wang et al., 2019a). Pseudomonas aeruginosa N17-1 degraded AFB1, AFB2, and AFM1 by 82.8%, 46.8%, and 31.9% after incubation in Nutrient Broth (NB) medium at 37 °C for 72 h with maximum degradation at 55 °C and after the addition of manganese (Mn2+) and copper (Cu2+) as activators (Sangare et al., 2014).
Pseudomonas aeruginosa N17-1 degraded AFB1, AFB2, and AFM1 by 82.8%, 46.8%, and 31.9% after incubation in Nutrient Broth (NB) medium at 37 °C for 72 h with maximum degradation at 55 °C and after the addition of manganese (Mn2+) and copper (Cu2+) as activators (Sangare et al., 2014).
FUNGI
 Aspergillus niger RAF106 effectively detoxified AFB1 to compounds with low or no mutagenicity in environments with pH values of 4-10 and temperatures of 25-45 °C (Fang et al., 2020).
Aspergillus niger RAF106 effectively detoxified AFB1 to compounds with low or no mutagenicity in environments with pH values of 4-10 and temperatures of 25-45 °C (Fang et al., 2020). A. niger FS10 degraded AFB1 by 98% into a less toxic compound after 12 h of incubation (Fang et al., 2020).
A. niger FS10 degraded AFB1 by 98% into a less toxic compound after 12 h of incubation (Fang et al., 2020).
Bioremediation of aflatoxins through enzymatic degradation
 Aflatoxins are degraded by a Mycobacterium smegmatis reductase enzyme that can utilize the deazaflavin cofactor F420H2 and catalyze the reduction of the α,β-unsaturated ester moiety of aflatoxins, activating the molecules for spontaneous hydrolysis and detoxification (Taylor et al., 2010).
Aflatoxins are degraded by a Mycobacterium smegmatis reductase enzyme that can utilize the deazaflavin cofactor F420H2 and catalyze the reduction of the α,β-unsaturated ester moiety of aflatoxins, activating the molecules for spontaneous hydrolysis and detoxification (Taylor et al., 2010). Manganese peroxidase produced by the white rot fungus Phanerochaete sordida YK-624 was able to detoxify AFB1 by an oxidation reaction consisting of firstly oxidizing to AFB1-8,9-epoxide by MnP and then hydrolyzing to AFB1– 8,9-dihydrodio (Wang et al., 2011).
Manganese peroxidase produced by the white rot fungus Phanerochaete sordida YK-624 was able to detoxify AFB1 by an oxidation reaction consisting of firstly oxidizing to AFB1-8,9-epoxide by MnP and then hydrolyzing to AFB1– 8,9-dihydrodio (Wang et al., 2011). The treatment of contaminated maize with culture filtrates of Trametes versicolor containing ligninolytic enzymes significantly reduced the content of AFB1 (Scarpari et al., 2014).
The treatment of contaminated maize with culture filtrates of Trametes versicolor containing ligninolytic enzymes significantly reduced the content of AFB1 (Scarpari et al., 2014). Manganese peroxidase, which can initiate oxidative lignin depolymerization in lignocellulose-degrading fungi lignocellulose Irpex lacteus, Phanerochaete chrysosporium, Ceriporiopsis subvermispora y Nematoloma forward, was able to degrade AFB1, ZEN, DON and FB1 (Wang et al., 2019d).
Manganese peroxidase, which can initiate oxidative lignin depolymerization in lignocellulose-degrading fungi lignocellulose Irpex lacteus, Phanerochaete chrysosporium, Ceriporiopsis subvermispora y Nematoloma forward, was able to degrade AFB1, ZEN, DON and FB1 (Wang et al., 2019d). The ability of C30 laccase from S. cerevisiae to degrade four aflatoxins (AFB1, AFB2, AFG1 and AFG2), showing that the binding capacity of aflatoxins to the enzyme was AFB1>AFG2>AFG1>AFB2.
The ability of C30 laccase from S. cerevisiae to degrade four aflatoxins (AFB1, AFB2, AFG1 and AFG2), showing that the binding capacity of aflatoxins to the enzyme was AFB1>AFG2>AFG1>AFB2.-
It was also demonstrated that aflatoxins may interact near the T1 copper center of the enzyme through H-bonds and hydrophobic interactions with amino acid residues. Furthermore, laccase was able to degrade all aflatoxins, and the degradation activity rose as incubation time increased from 12 to 60 h, with maximum degradation for AFB1, AFB2, AFG1, and AFG2 of 90.33%, 74.23%, 85.24%, and 87.58%, respectively, at 30 °C in 0.1 M PBS and pH 5.7 (Liu et al., 2020).
 A laccase enzyme from the fungus Pleurotus pulmonarius degraded AFB1 under optimal conditions of pH 7 and a temperature of 37 °C (Song et al., 2021).
A laccase enzyme from the fungus Pleurotus pulmonarius degraded AFB1 under optimal conditions of pH 7 and a temperature of 37 °C (Song et al., 2021). A laccase from Bacillus amyloliquefaciens B10 was separated and found to degrade 79.3% of AFB1 within 36 h under an optimum temperature of 40 °C and pH 6–8 with the addition of magnesium (Mg2+) and dimethyl sulfoxide (DMSO) as reaction enhancers (Xiong et al., 2022).
A laccase from Bacillus amyloliquefaciens B10 was separated and found to degrade 79.3% of AFB1 within 36 h under an optimum temperature of 40 °C and pH 6–8 with the addition of magnesium (Mg2+) and dimethyl sulfoxide (DMSO) as reaction enhancers (Xiong et al., 2022). A laccase gene was cloned from Bacillus vallismortis fmb-103 and expressed in heterologous host E. coli after codon optimization and its degradation of AFB1 was investigated and found to be improved with an increase in incubation pH and temperature and exceeded 60% at pH 7 and 37 °C (Bian et al., 2022).
A laccase gene was cloned from Bacillus vallismortis fmb-103 and expressed in heterologous host E. coli after codon optimization and its degradation of AFB1 was investigated and found to be improved with an increase in incubation pH and temperature and exceeded 60% at pH 7 and 37 °C (Bian et al., 2022). An AFB1-degrading enzyme produced by Pseudomonas aeruginosa degraded AFB1 under optimal conditions of 65°C and pH 6 with the presence of Cu2+ and iron (Fe3+) (Song et al., 2019).
An AFB1-degrading enzyme produced by Pseudomonas aeruginosa degraded AFB1 under optimal conditions of 65°C and pH 6 with the presence of Cu2+ and iron (Fe3+) (Song et al., 2019). AFB1-degrading proteins or enzymes in the intracellular extracts of A. niger RAF106 induced 80% degradation of AFB1 over 24 h of incubation (Fang et al., 2020).
AFB1-degrading proteins or enzymes in the intracellular extracts of A. niger RAF106 induced 80% degradation of AFB1 over 24 h of incubation (Fang et al., 2020). Aflatoxin-oxidase produced by Armillariella tabescens, a Chinese edible and medicinal fungus, was effective in AFB1 degradation by acting on the bisfuran ring of AFB1 which is considered the key toxic structure (Cao et al., 2011).
Aflatoxin-oxidase produced by Armillariella tabescens, a Chinese edible and medicinal fungus, was effective in AFB1 degradation by acting on the bisfuran ring of AFB1 which is considered the key toxic structure (Cao et al., 2011).
OCHRATOXINS
Ochratoxins (OTs) are extremely toxic mycotoxins.
Ochratoxin A (OTA), the most frequent type found in food and feedstuff is produced mainly by Aspergillus ochraceus, whereas ochratoxin B and C are produced by other Aspergillus and Penicillium species (Marroquín-Cardona et al., 2014).
OTA is the most toxic and prevalent in the ochratoxin family (Heussner y Bingle, 2015).
It is nephrotoxic (Khoi et al., 2021), genotoxic, cytotoxic, teratogenic (Hibi et al., 2011), carcinogenic (Pfohl-Leszkowicz y Manderville, 2012), mutagenic, immunotoxic, and myelotoxic (Marin and Taranu, 2015).
Bioremediation of OTA by microbial adsorption
Various microorganisms have been reported to be able to remove OTA through adsorption. The ability of microorganisms to adsorb OTA is
 Microbial strains (their status is either viable or inviable) (Bejaoui et al., 2004; Piotrowska, 2014).
Microbial strains (their status is either viable or inviable) (Bejaoui et al., 2004; Piotrowska, 2014). Culture conditions, including pH and temperature (Gil-Serna et al., 2011).
Culture conditions, including pH and temperature (Gil-Serna et al., 2011).
LACTIC ACID BACTERIA
 In a recent study, detoxification of OTA was investigated using single and co-culture of Lactobacillus acidophilus and L. rhamnosus in various modes, including viable, heated, and ultrasound-inactivated cells, as well as a combination of these conditions and the results, were different depending on cell wall structure, hydrophobicity, and toxin binding capacity of single and co-culture of lactic acid bacteria (Abedi et al., 2022).
In a recent study, detoxification of OTA was investigated using single and co-culture of Lactobacillus acidophilus and L. rhamnosus in various modes, including viable, heated, and ultrasound-inactivated cells, as well as a combination of these conditions and the results, were different depending on cell wall structure, hydrophobicity, and toxin binding capacity of single and co-culture of lactic acid bacteria (Abedi et al., 2022).
 The binding abilities of three thermally active or inactivated lactic acid bacteria species, Lactobacillus brevis, L. plantarum, and Lactobacillus sanfranciscensis were investigated, and it was reported that the binding by inactivated bacterial cells was higher than viable cells, and OTA adsorption to the cell wall was attributed to hydrophobic and electrostatic interactions (Piotrowska, 2014).
The binding abilities of three thermally active or inactivated lactic acid bacteria species, Lactobacillus brevis, L. plantarum, and Lactobacillus sanfranciscensis were investigated, and it was reported that the binding by inactivated bacterial cells was higher than viable cells, and OTA adsorption to the cell wall was attributed to hydrophobic and electrostatic interactions (Piotrowska, 2014). 54 lactic acid bacteria were isolated from stored wheat samples and studied for their ability to remove OTA in a liquid medium and the results were dependent on temperature, pH, and bacterial biomass on mixed cultures with a high percentage of OTA reduction obtained by L. plantarum and Lactobacillus graminis (more than 97%) followed by Pediococcus pentosaceus (more than 81.5%) (Belkacem-Hanfi et al., 2014).
54 lactic acid bacteria were isolated from stored wheat samples and studied for their ability to remove OTA in a liquid medium and the results were dependent on temperature, pH, and bacterial biomass on mixed cultures with a high percentage of OTA reduction obtained by L. plantarum and Lactobacillus graminis (more than 97%) followed by Pediococcus pentosaceus (more than 81.5%) (Belkacem-Hanfi et al., 2014).
 P. pentosaceus (RG7B) removed 84% of OTA in MRS media, while its binding capacity in PBS media was only 25% (Taroub et al., 2019).
P. pentosaceus (RG7B) removed 84% of OTA in MRS media, while its binding capacity in PBS media was only 25% (Taroub et al., 2019). Fourteen strains isolated from LAB and Bifidobacterium were tested to evaluate their ability to bind OTA in PBs contaminated by 10 ppb OTA. These tested strains reduced 80.4 to 81.4% OTA at 37 ⁰C in 36 h (Fouad et al., 2021).
Fourteen strains isolated from LAB and Bifidobacterium were tested to evaluate their ability to bind OTA in PBs contaminated by 10 ppb OTA. These tested strains reduced 80.4 to 81.4% OTA at 37 ⁰C in 36 h (Fouad et al., 2021).
YEASTS
 Bejaoui et al. (2004) reported that heat and acid-treated yeasts could adsorb 90.8% and 73% of OTA, respectively, while viable cells bound up to 35% of OTA.
Bejaoui et al. (2004) reported that heat and acid-treated yeasts could adsorb 90.8% and 73% of OTA, respectively, while viable cells bound up to 35% of OTA.-
In the same study, the adsorption capacity under heat or acid treatment was attributed to their ability to alter the surface features of microbial cells, such as the enlargement of pore size, the denaturation of proteins, and the formation of Maillard reaction products between polysaccharides and peptides.
 Four S. cerevisiae strains were assessed for their OTA removal capabilities and the results showed that OTA adsorbing efficiencies varied according to the yeast strains and OTA concentrations. Moreover, there was a correlation between the mycotoxin removal capability and cell wall amount, suggesting the yeast-mycotoxin interaction is more of an adsorption type (Armando et al., 2012).
Four S. cerevisiae strains were assessed for their OTA removal capabilities and the results showed that OTA adsorbing efficiencies varied according to the yeast strains and OTA concentrations. Moreover, there was a correlation between the mycotoxin removal capability and cell wall amount, suggesting the yeast-mycotoxin interaction is more of an adsorption type (Armando et al., 2012).
 S. cerevisiae (both Syrena LOCK 0201 and Malaga LOCK 0173 strains) decreased the amount of OTA.
S. cerevisiae (both Syrena LOCK 0201 and Malaga LOCK 0173 strains) decreased the amount of OTA.-
Strain Malaga LOCK 0173 was able to remove 82.8% and 10.7% from grape and blackcurrant mediums, respectively, while Syrena LOCK 0201 strain’s binding capacity was 85.1% for grape and 65.2% for blackcurrant media (Piotrowska et al., 2013).
 Two strains of S. cerevisiae decreased the level of OTA by 70% in 3 days at 30°C in the presence of 250 g l−1 of sugars and with diammonium phosphate (Petruzzi et al., 2014).
Two strains of S. cerevisiae decreased the level of OTA by 70% in 3 days at 30°C in the presence of 250 g l−1 of sugars and with diammonium phosphate (Petruzzi et al., 2014).

OTHER MICROORGANISMS
 A Bacillus megaterium strain JSW-B1 was reported to bind 62.6% OTA in a liquid culture medium within 72 h and heat treatment remarkably increased the OTA binding ability of the cells, indicating the OTA removal activity of this strain mainly resulted from the adsorption to bacterial cells (Shang et al., 2019).
A Bacillus megaterium strain JSW-B1 was reported to bind 62.6% OTA in a liquid culture medium within 72 h and heat treatment remarkably increased the OTA binding ability of the cells, indicating the OTA removal activity of this strain mainly resulted from the adsorption to bacterial cells (Shang et al., 2019).
 The use of Bacillus subtilis immobilized in edible sodium alginate beads (pH 3 and 25% sodium chloride, NaCl) was highly effective in the reduction of OTA in red wine where it reduced 90% of OTA from culture and when these beads were added to red wine spiked with OTA, it reduced OTA by 78.50 ± 0.49% after 1 h (Shukla et al., 2020).
The use of Bacillus subtilis immobilized in edible sodium alginate beads (pH 3 and 25% sodium chloride, NaCl) was highly effective in the reduction of OTA in red wine where it reduced 90% of OTA from culture and when these beads were added to red wine spiked with OTA, it reduced OTA by 78.50 ± 0.49% after 1 h (Shukla et al., 2020).
![]() Recently, OTA adsorption to microorganisms has been proved to be genetically controlled. For example:
Recently, OTA adsorption to microorganisms has been proved to be genetically controlled. For example:
 L. delbrueckii NRRL B-1024 was mutagenized using ethyl methanesulfonate (EMS) to induce genetic variations to improve OTA biosorption. Seven of 19 mutants exhibited OTA biosorption higher than the wild-type strain (Khattab et al., 2018).
L. delbrueckii NRRL B-1024 was mutagenized using ethyl methanesulfonate (EMS) to induce genetic variations to improve OTA biosorption. Seven of 19 mutants exhibited OTA biosorption higher than the wild-type strain (Khattab et al., 2018). Twelve S. cerevisiae strains were used to obtain intraspecific hybrids and five of the 27 validated hybrids exhibited higher OTA but lower phenolic compounds adsorption activity as compared to their parents (Caridi et al., 2020).
Twelve S. cerevisiae strains were used to obtain intraspecific hybrids and five of the 27 validated hybrids exhibited higher OTA but lower phenolic compounds adsorption activity as compared to their parents (Caridi et al., 2020).
Bioremediation of OTA by microbial degradation
La degradación microbiana de la OTA se produce a través de dos reacciones catalizadas por enzimas:
 Hydrolysis of the amide bond between the isocoumarin residue and phenylalanine.
Hydrolysis of the amide bond between the isocoumarin residue and phenylalanine. Hydrolysis of the lactone ring.
Hydrolysis of the lactone ring.
Among the reported degradation products, OTα is considered non-toxic or less toxic (Bittner et al., 2015; Haq et al., 2016) and several bacterial and fungal strains have proven their capacity to degrade OTA with or without producing OTα.
BACTERIA
 B. licheniformis Sl-1 degraded OTA in liquid media where autoclaved cells bound 80% OTA while viable cells 60% OTA after 24 h aerobic incubation.
B. licheniformis Sl-1 degraded OTA in liquid media where autoclaved cells bound 80% OTA while viable cells 60% OTA after 24 h aerobic incubation.-
The cell-free supernatant of Sl-1 degraded OTA with a 98% degradation rate after 24 h of aerobic incubation. No degradation products were produced and, when applied to mouldy maize samples, the OTA degradation rate was 35% (Shi et al., 2013).
 OTA was degraded by both viable and autoclaved cells of B. subtilis CW 14 by more than 60% and it was degraded by the cell-free supernatant of CW 14 by a 97.6% degradation rate after 24 h of incubation at 30 °C with no produced degradation products. In addition, using culture inoculation degraded 47.1% of OTA in contaminated maize samples (Shi et al., 2014).
OTA was degraded by both viable and autoclaved cells of B. subtilis CW 14 by more than 60% and it was degraded by the cell-free supernatant of CW 14 by a 97.6% degradation rate after 24 h of incubation at 30 °C with no produced degradation products. In addition, using culture inoculation degraded 47.1% of OTA in contaminated maize samples (Shi et al., 2014). B. velezensis E2 degraded 96.1% of OTA within 48 h by enzymatic transformation and alkaline hydrolysis producing open ochratoxin α (OP-OTα) as a degradation product (Zhang et al., 2022).
B. velezensis E2 degraded 96.1% of OTA within 48 h by enzymatic transformation and alkaline hydrolysis producing open ochratoxin α (OP-OTα) as a degradation product (Zhang et al., 2022).
FUNGI
 Aspergillus tubingensis M036 and M074 strains degraded OTA by more than 95% after 14 days, producing ochratoxin α.
Aspergillus tubingensis M036 and M074 strains degraded OTA by more than 95% after 14 days, producing ochratoxin α.-
Moreover, crude enzymes from the cultures of both strains led to OTA biodegradation by 97.5% and 91.3% at pH 5 and 80.3% and 75.3% at pH 7, respectively, after 24 h (Cho et al., 2016).
 A niger W-35 degraded OTA in vitro and in commercial feeds after incubation at 37 °C for 12 h by 78% and 37%, respectively, with a production of OTα.
A niger W-35 degraded OTA in vitro and in commercial feeds after incubation at 37 °C for 12 h by 78% and 37%, respectively, with a production of OTα.-
Furthermore, an enzyme specific for OTA degradation (ochratoxinase, OTase) obtained from W-35 was successfully expressed in E. coli BL21 and degraded OTA at a rate of 85.1% for 12 h (Zhao et al., 2020).
 A novel Aspergillus oryzae strain was able to degrade 94% of OTA after 72 h of incubation (Xiong et al., 2021). In addition, Aspergillus calcoaceticus strain 396.1 and Acinetobacter sp. strain neg1 were able to convert 82% and 91% OTA into OTα in 6 days of incubation at 24 °C (De Bellis et al., 2015).
A novel Aspergillus oryzae strain was able to degrade 94% of OTA after 72 h of incubation (Xiong et al., 2021). In addition, Aspergillus calcoaceticus strain 396.1 and Acinetobacter sp. strain neg1 were able to convert 82% and 91% OTA into OTα in 6 days of incubation at 24 °C (De Bellis et al., 2015). Lactobacillus johnsonii CECT 289, L. rhamnosus CECT 278T and L. plantarum CECT 749 had 97.4 %, 97%, and 95% degradation values of OTA into OTα and phenylalanine (Phe) (Luz et al., 2018).
Lactobacillus johnsonii CECT 289, L. rhamnosus CECT 278T and L. plantarum CECT 749 had 97.4 %, 97%, and 95% degradation values of OTA into OTα and phenylalanine (Phe) (Luz et al., 2018).
Bioremediation of OTA by enzymatic degradation
 Carboxypeptidase, a hydrolysed enzyme related to the enzymatic conversion of OTA, was cloned from the B. amyloliquefaciens ASAG1 genome using the E. coli Expression System and it was reported to have high OTA degrading activities (Chang et al., 2015).
Carboxypeptidase, a hydrolysed enzyme related to the enzymatic conversion of OTA, was cloned from the B. amyloliquefaciens ASAG1 genome using the E. coli Expression System and it was reported to have high OTA degrading activities (Chang et al., 2015). Carboxypeptidase gene dacA from B. subtilis CW14 expressed in Escherichia coli, contributed to degrade OTA by 71.3% (Xu et al., 2021).
Carboxypeptidase gene dacA from B. subtilis CW14 expressed in Escherichia coli, contributed to degrade OTA by 71.3% (Xu et al., 2021). OTA was degraded under the activity of carboxypeptidase PJ_1540 produced by Acinetobacter sp. neg1, ITEM 17016 (Liuzzi et al., 2017).
OTA was degraded under the activity of carboxypeptidase PJ_1540 produced by Acinetobacter sp. neg1, ITEM 17016 (Liuzzi et al., 2017).
 Another study investigated the capacity of laccase enzyme produced by Pleurotus eryngii and laccase-mediator systems (LMSs) to degrade OTA, and it was found to degrade 27% OTA (Loi et al., 2018).
Another study investigated the capacity of laccase enzyme produced by Pleurotus eryngii and laccase-mediator systems (LMSs) to degrade OTA, and it was found to degrade 27% OTA (Loi et al., 2018). Another enzyme reported to be capable of OTA degradation is ochratoxinase from A. niger.
Another enzyme reported to be capable of OTA degradation is ochratoxinase from A. niger.-
In a study by Dobritzsch et al. (2014). A putative amidase gene encoding a 480 amino acid polypeptide was cloned and homologously expressed in A. niger. The recombinant protein is believed to be more efficient in ochratoxin A hydrolysis than carboxypeptidase enzyme.
ZEARALENONE
Zearalenone (ZEN) is a non-steroidal estrogenic mycotoxin produced mainly by Fusarium graminearum and, to a minor extent, by Fusarium culmorum, Fusarium cerealis or Fusarium equiseti (De Boevre et al., 2012; Taheur et al., 2017).
ZEN occurs naturally in maize, barley, oats, wheat, rice, sorghum, rye, and other major commodities when humidity and temperature conditions are suitable (Golge y Kabak, 2020).
It can activate estrogenic receptors, resulting in reproductive disorders in animals (Gupta, 2012) and fish species (Schwartz et al., 2013; Woźny et al., 2020).
ZEN and some of its derivatives can also induce genotoxic effects by induction of DNA adducts, DNA fragmentation, apoptosis, micronuclei, and chromosome aberrations (Gao et al., 2013).
Bioremediation of ZEN by microbial adsorption
ZEN can be adsorbed to microbial cell walls via their proteins, lipids, and carbohydrates (mannose, peptidoglycans, glucans).
The adsorption mechanisms include ionic interactions, hydrogen bonding, and hydrophobic interactions (Wang et al., 2019b) and they depend on many factors, including:
 Microbial strains: the presence of carbohydrates in their cell wall and its thickness
Microbial strains: the presence of carbohydrates in their cell wall and its thickness Culture conditions: pH and temperature, incubation time, and the addition of physical and chemical enhancers
Culture conditions: pH and temperature, incubation time, and the addition of physical and chemical enhancers

SACCHAROMYCES
 Adding 0.2% yeast cell wall adsorbent to ZEN-contaminated corn effectively alleviated piglet reproductive toxicity (Shang et al., 2016).
Adding 0.2% yeast cell wall adsorbent to ZEN-contaminated corn effectively alleviated piglet reproductive toxicity (Shang et al., 2016). 10 strains of S. cerevisiae and 5 of Saccharomyces pastorianus could adsorb 30-70% ZEN depending on strain in contaminated wort after a 96 h fermentation period (Wall-Martínez et al., 2019).
10 strains of S. cerevisiae and 5 of Saccharomyces pastorianus could adsorb 30-70% ZEN depending on strain in contaminated wort after a 96 h fermentation period (Wall-Martínez et al., 2019). The ability of beer fermentation residue containing S. cerevisiae to bind ZEN was investigated at different pH levels and it was found that 75.1% and 77.5% of ZEN were removed at pH 3 and 6.5, respectively (Campagnollo et al., 2015).
The ability of beer fermentation residue containing S. cerevisiae to bind ZEN was investigated at different pH levels and it was found that 75.1% and 77.5% of ZEN were removed at pH 3 and 6.5, respectively (Campagnollo et al., 2015).
 The ability of S. cerevisiae to decrease the ZEN content can be influenced by physical or chemical pre-treatment.
The ability of S. cerevisiae to decrease the ZEN content can be influenced by physical or chemical pre-treatment.-
For instance, the maximum adsorption capacity of S. cerevisiae incubated under stress-free conditions was 36.5%. In comparison, incubation under stress conditions (induced by silver nanoparticles and lyophilization) increased the maximum sorption value to 57.0% (Rogowska et al., 2019).
 The cell diameter/cell wall thickness relation showed a correlation between the cell wall amount and ZEN removal ability, and there was a significant increase in ZEN binding after exposure to gastrointestinal conditions (Armando et al., 2012).
The cell diameter/cell wall thickness relation showed a correlation between the cell wall amount and ZEN removal ability, and there was a significant increase in ZEN binding after exposure to gastrointestinal conditions (Armando et al., 2012).-
It has been suggested that S. cerevisiae may bind mycotoxins to their cell wall via their contained functional carbohydrates (glucomannan polymers).
 It was reported that the cell wall of S. cerevisiae could bind ZEN and found that The β(1,3)-d-glucan and β(1,6)- d-glucan were the most favorable structures for zearalenone adsorption (Yiannikouris et al., 2004).
It was reported that the cell wall of S. cerevisiae could bind ZEN and found that The β(1,3)-d-glucan and β(1,6)- d-glucan were the most favorable structures for zearalenone adsorption (Yiannikouris et al., 2004). In a more recent study, S. cerevisiae NRRL Y-1089 and S. cerevisiae NRRL Y-1089 removed ZEN by 94.80% and 91.80%, respectively, and the removal of ZEN was decreased by increasing the incubation period (Hathout y Abdel-Nasser, 2022).
In a more recent study, S. cerevisiae NRRL Y-1089 and S. cerevisiae NRRL Y-1089 removed ZEN by 94.80% and 91.80%, respectively, and the removal of ZEN was decreased by increasing the incubation period (Hathout y Abdel-Nasser, 2022).
LACTIC ACID BACTERIA
 Eight Lactobacillus pentosus strains were evaluated for their ability to adsorb ZEN from a sodium acetate buffer solution. The adsorption capacity was 83.17%, 82.78% and 81.69% for strains JM0812, UM054 and UM055, respectively (Sangsila et al., 2016).
Eight Lactobacillus pentosus strains were evaluated for their ability to adsorb ZEN from a sodium acetate buffer solution. The adsorption capacity was 83.17%, 82.78% and 81.69% for strains JM0812, UM054 and UM055, respectively (Sangsila et al., 2016). L. rhamnosus strains GG and LC705 could bind 55% of ZEN and α-ZOL, and it was reported that both toxins might share the same binding site on the bacterial surface (El-Nezami et al., 2002).
L. rhamnosus strains GG and LC705 could bind 55% of ZEN and α-ZOL, and it was reported that both toxins might share the same binding site on the bacterial surface (El-Nezami et al., 2002). In a more recent study, Lactococcus lactis and Bifidobacterium sp were able to adsorb ZEN by 90 % and 88%, respectively, via binding to deprotonated carboxyl groups (mainly from asparagine and glutamine) from LAB proteins and peptidoglycan (Król et al., 2018).
In a more recent study, Lactococcus lactis and Bifidobacterium sp were able to adsorb ZEN by 90 % and 88%, respectively, via binding to deprotonated carboxyl groups (mainly from asparagine and glutamine) from LAB proteins and peptidoglycan (Król et al., 2018).
 L. rhamnosus and other species could absorb from 40% to 68.2% of ZEN, and these strains could form a permanent bond with ZEN, which inspired the design of a new freeze-dried powder based on LAB stains for ZEN detoxification (Vega et al., 2017).
L. rhamnosus and other species could absorb from 40% to 68.2% of ZEN, and these strains could form a permanent bond with ZEN, which inspired the design of a new freeze-dried powder based on LAB stains for ZEN detoxification (Vega et al., 2017). ZEN adsorption by L. lactis was nonhomogeneous where it was expressed with initial rapid stage with about 90% of ZEN biosorption and with a much slower second step while the adsorption by Bifidobacterium sp was homogeneous by 88% (Król et al., 2017).
ZEN adsorption by L. lactis was nonhomogeneous where it was expressed with initial rapid stage with about 90% of ZEN biosorption and with a much slower second step while the adsorption by Bifidobacterium sp was homogeneous by 88% (Król et al., 2017). Other LAB strains were found to adsorb multiple mycotoxins. Lactobacillus kefiri, Kazachstania servazzii and Acetobacter syzygii were reported to adsorb ZEN, AFB1 and OTA where L. kefiri KFLM3 was the most active, adsorbing 80 to 100% of the mycotoxins when cultivated in milk while the strain K. servazzii KFGY7 bound 65, 69 and 67% for AFB1, OTA and ZEN, respectively (Taheur et al., 2017).
Other LAB strains were found to adsorb multiple mycotoxins. Lactobacillus kefiri, Kazachstania servazzii and Acetobacter syzygii were reported to adsorb ZEN, AFB1 and OTA where L. kefiri KFLM3 was the most active, adsorbing 80 to 100% of the mycotoxins when cultivated in milk while the strain K. servazzii KFGY7 bound 65, 69 and 67% for AFB1, OTA and ZEN, respectively (Taheur et al., 2017). In the same line, Streptococcus and Enterococcus species were capable of binding up to 33, 49, 24 and 62% of DON, ZEN, FB1 and FB2, respectively (Niderkorn et al., 2007).
In the same line, Streptococcus and Enterococcus species were capable of binding up to 33, 49, 24 and 62% of DON, ZEN, FB1 and FB2, respectively (Niderkorn et al., 2007).
Bioremediation of ZEN by microbial degradation
 ZEN was completely degraded by S. cerevisiae after 48 h of incubation and it was suggested that the mechanisms involved in the degradation were the production of intracellular and extracellular enzymes and some functional proteins produced by S. cerevisiae (Zhang et al., 2016).
ZEN was completely degraded by S. cerevisiae after 48 h of incubation and it was suggested that the mechanisms involved in the degradation were the production of intracellular and extracellular enzymes and some functional proteins produced by S. cerevisiae (Zhang et al., 2016). Several S. cerevisiae strains were able to degrade more than 90% of ZEN from the culture medium in two days with the production of less toxic α- and β-zearalenol (α-ZOL 53% and β-ZOL 8%) as end products suggesting that the elimination was mainly due to ZEN biotransformation rather than to its adsorption to yeast cells walls (Keller et al., 2015).
Several S. cerevisiae strains were able to degrade more than 90% of ZEN from the culture medium in two days with the production of less toxic α- and β-zearalenol (α-ZOL 53% and β-ZOL 8%) as end products suggesting that the elimination was mainly due to ZEN biotransformation rather than to its adsorption to yeast cells walls (Keller et al., 2015).
 B. subtilis ANSB01G degraded 84.58, 66.34 and 83.04% of ZEN in naturally contaminated maize, dried distillers’ grains with solubles, and swine complete feed, respectively.
B. subtilis ANSB01G degraded 84.58, 66.34 and 83.04% of ZEN in naturally contaminated maize, dried distillers’ grains with solubles, and swine complete feed, respectively.-
The degradation was believed to be enzymatic because treatments with heating or adding proteinase K significantly reduced the rate of ZEN degradation in the culture supernatant (Lei et al., 2014).
 B. amyloliquefaciens ZDS-1 degraded ZEN in a culture medium and wheat through enzymatic degradation with no production of ZEN derivatives (Xu et al., 2016).
B. amyloliquefaciens ZDS-1 degraded ZEN in a culture medium and wheat through enzymatic degradation with no production of ZEN derivatives (Xu et al., 2016). B. amyloliquefaciens LN decreased ZEN concentration by 92% after 36 h of incubation and it was reported that B. amyloliquefaciens LN not only possessed ZEN adsorption capacity but also exhibited the ability to degrade ZEN. In addition, B. amyloliquefaciens LN was non-hemolytic, non-enterotoxin producing (Lee et al., 2017).
B. amyloliquefaciens LN decreased ZEN concentration by 92% after 36 h of incubation and it was reported that B. amyloliquefaciens LN not only possessed ZEN adsorption capacity but also exhibited the ability to degrade ZEN. In addition, B. amyloliquefaciens LN was non-hemolytic, non-enterotoxin producing (Lee et al., 2017).
 Numerous Bacillus strains were able to degrade ZEN, including B. licheniformis which decreased more than 98% of ZEN after 36 h of incubation through non-hemolytic, non-enterotoxin-producing mechanisms by producing high levels of extracellular xylanase, cellulase, and protease activities (Yi et al., 2011).
Numerous Bacillus strains were able to degrade ZEN, including B. licheniformis which decreased more than 98% of ZEN after 36 h of incubation through non-hemolytic, non-enterotoxin-producing mechanisms by producing high levels of extracellular xylanase, cellulase, and protease activities (Yi et al., 2011). ZEN degradation rates of B. subtilis and B. natto were 100% and 87% within 48 h, respectively; the two taxa have high degradation activity for ZEN in beer, corn flour, and soy flour (Ju et al., 2019).
ZEN degradation rates of B. subtilis and B. natto were 100% and 87% within 48 h, respectively; the two taxa have high degradation activity for ZEN in beer, corn flour, and soy flour (Ju et al., 2019).
Biodegradation of ZEN by microbial enzymes
Three main types of biological enzymes can degrade ZEN: laccase, lactone hydrolase, and peroxidase.
They attack three different sites on the ZEN molecule:
- 1. The first position is the ester bond of the lactone ring, and if the lactone ring can be opened, the resulting molecule loses its estrogen-mimicking properties.
- 2. The second reaction site is on the long lactone ring, especially the C-C bond near the carbonyl group.
- 3. The third one involves opening up the aromatic ring as a way to degrade ZEN.
(Kriszt et al., 2012; Wu et al., 2021)
LACTONE HYDROLASES
Lactone hydrolases are the most studied for ZEN degradation.
 The lactone hydrolase ZHD101 was extracted from the mycoparasitic fungus Clonostachys rosea and degraded ZEN into less toxic products (Takahashi-Ando et al., 2002).
The lactone hydrolase ZHD101 was extracted from the mycoparasitic fungus Clonostachys rosea and degraded ZEN into less toxic products (Takahashi-Ando et al., 2002). Gliocladium roseum was found to produce lactone hydrolase that can break and hydrolyze the lactone bond of ZEN to form non-toxic ZEN derivatives (Utermark y Karlovsky, 2007).
Gliocladium roseum was found to produce lactone hydrolase that can break and hydrolyze the lactone bond of ZEN to form non-toxic ZEN derivatives (Utermark y Karlovsky, 2007). Bi et al. (2018) reported that the ZEN lactonase from Neurospora crassa is encoded by the zenc gene and over-expressed in Pichia pastoris.
Bi et al. (2018) reported that the ZEN lactonase from Neurospora crassa is encoded by the zenc gene and over-expressed in Pichia pastoris. Subsequently, the purified zenc enzyme was added to the distiller’s dried grains with solubles (DDGS), maize byproducts, and corn bran, and the concentration of ZEN was decreased by 71%, 89%, and 95%, respectively.
Subsequently, the purified zenc enzyme was added to the distiller’s dried grains with solubles (DDGS), maize byproducts, and corn bran, and the concentration of ZEN was decreased by 71%, 89%, and 95%, respectively.
LACCASE
 Laccase enzyme was isolated from fungal species T. versicolor and it was found to decrease the ZEN content in a liquid medium by 81.7 % after 4 h incubation at its optimum temperature (Banu et al., 2013).
Laccase enzyme was isolated from fungal species T. versicolor and it was found to decrease the ZEN content in a liquid medium by 81.7 % after 4 h incubation at its optimum temperature (Banu et al., 2013). CotA laccase from B. subtilis (BsCotA) combined with the chemical compound methyl syringate degraded ZEN by 100% (Wang et al., 2019c).
CotA laccase from B. subtilis (BsCotA) combined with the chemical compound methyl syringate degraded ZEN by 100% (Wang et al., 2019c). The ability of laccase (LC) from P. eryngii and laccase-mediator systems (LMSs) to degrade ZEN was investigated, and it was found that ZEN was degraded by 100% (Loi et al., 2018).
The ability of laccase (LC) from P. eryngii and laccase-mediator systems (LMSs) to degrade ZEN was investigated, and it was found that ZEN was degraded by 100% (Loi et al., 2018). A laccase enzyme from the fungus Pleurotus pulmonarius was able to degrade ZEN under optimal conditions of pH (4–8) and temperature (40-60 ◦C) (Song et al., 2021).
A laccase enzyme from the fungus Pleurotus pulmonarius was able to degrade ZEN under optimal conditions of pH (4–8) and temperature (40-60 ◦C) (Song et al., 2021).
PEROXIDASE
Other studies have shown the capacity of peroxidase enzyme to degrade ZEN.
-
 The extracellular extracts of the Acinetobacter genus completely degraded zearalenone strain SM04 in M1 medium after 12 h of incubation (Yu et al., 2011b).
The extracellular extracts of the Acinetobacter genus completely degraded zearalenone strain SM04 in M1 medium after 12 h of incubation (Yu et al., 2011b).  Enzymes from Acinetobacter sp were extracted and ZEN was degraded to smaller non-estrogenic products.
Enzymes from Acinetobacter sp were extracted and ZEN was degraded to smaller non-estrogenic products.-
The enzyme was identified as peroxiredoxin, and hydrogen peroxide oxidized ZEN and its products by the catalysis of peroxiredoxin (Yu et al., 2011a).
 Wang et al. (2019d) reported that eight manganese peroxidases from different lignocellulose-degrading fungi could degrade 90.2–94.3% of ZEN in the presence of malonate, whereby free radicals play an important role.
Wang et al. (2019d) reported that eight manganese peroxidases from different lignocellulose-degrading fungi could degrade 90.2–94.3% of ZEN in the presence of malonate, whereby free radicals play an important role.

Conclusions
Bacteria, yeasts, and fungi can detoxify mycotoxins efficiently and specifically.
Microbial degradation decomposes toxins without leaving toxic residues for food and feed safety. Furthermore, microbial degradation and adsorption can convert several common mycotoxins into relatively safe substances, making food and feed safer.
Recent studies on microbial mycotoxins in food or feed have focused on removing single mycotoxins and applying strains in the lab.
This review proposes several future directions:
- 1. First, detoxified strains or food-degrading enzymes.
2. Second, to improve degradation and reduce mycotoxin stability, studying the combination of multiple strains and microbial degradation of mycotoxins.
REFERENCES
Abbès, S., Salah-Abbès, J.B., Sharafi, H., Jebali, R., Noghabi, K.A., Oueslati, R., 2013. Ability of Lactobacillus rhamnosus GAF01 to remove AFM1 in vitro and to counteract AFM1 immunotoxicity in vivo. Journal of Immunotoxicology 10, 279-286.
Abedi, E., Mousavifard, M., Hashemi, S.M.B., 2022. Ultrasound-Assisted Detoxification of Ochratoxin A: Comparative Study of Cell Wall Structure, Hydrophobicity, and Toxin Binding Capacity of Single and Co-culture Lactic Acid Bacteria. Food and Bioprocess Technology 15, 539-560.
Adejumo, T., Adejoro, D., 2014. Incidence of aflatoxins, fumonisins, trichothecenes and ochratoxins in Nigerian foods and possible intervention strategies. Food Science and Quality Management 31, 127-147.
Alberts, J., Gelderblom, W., Botha, A., Van Zyl, W., 2009. Degradation of aflatoxin B1 by fungal laccase enzymes. International journal of food microbiology 135, 47-52.
Alshannaq, A., Yu, J.-H., 2017. Occurrence, toxicity, and analysis of major mycotoxins in food. International journal of environmental research and public health 14, 632.
Anater, A., Manyes, L., Meca, G., Ferrer, E., Luciano, F.B., Pimpao, C.T., Font, G., 2016. Mycotoxins and their consequences in aquaculture: A review. Aquaculture 451, 1-10.
Armando, M., Pizzolitto, R., Dogi, C., Cristofolini, A., Merkis, C., Poloni, V., Dalcero, A., Cavaglieri, L.R., 2012. Adsorption of ochratoxin A and zearalenone by potential probiotic Saccharomyces cerevisiae strains and its relation with cell wall thickness. Journal of applied microbiology 113, 256-264.
Ashiq, S., 2015. Natural occurrence of mycotoxins in food and feed: Pakistan perspective. Comprehensive reviews in food science and food safety 14, 159-175.
Banu, I., Lupu, A., Aprodu, I., 2013. Degradation of zearalenone by laccase enzyme. Scientific Study & Research. Chemistry & Chemical Engineering, Biotechnology, Food Industry 14, 79.
Bejaoui, H., Mathieu, F., Taillandier, P., Lebrihi, A., 2004. Ochratoxin A removal in synthetic and natural grape juices by selected oenological Saccharomyces strains. Journal of applied microbiology 97, 1038-1044.
Belkacem-Hanfi, N., Fhoula, I., Semmar, N., Guesmi, A., Perraud-Gaime, I., Ouzari, H.-I., Boudabous, A., Roussos, S., 2014. Lactic acid bacteria against post-harvest moulds and ochratoxin A isolated from stored wheat. Biological Control 76, 52-59.
Bhatnagar, D., Payne, G.A., Cleveland, T.E., Robens, J.F. 2004. Mycotoxins: current issues in USA. In: Meeting the mycotoxin menace, 17-47.
Bi, K., Zhang, W., Xiao, Z., Zhang, D., 2018. Characterization, expression and application of a zearalenone degrading enzyme from Neurospora crassa. Amb Express 8, 1-10.
Bian, L., Zheng, M., Chang, T., Zhou, J., Zhang, C., 2022. Degradation of Aflatoxin B1 by recombinant laccase extracellular produced from Escherichia coli. Ecotoxicology and Environmental Safety 244, 114062.
Bittner, A., Cramer, B., Harrer, H., Humpf, H.-U., 2015. Structure elucidation and in vitro cytotoxicity of ochratoxin α amide, a new degradation product of ochratoxin A. Mycotoxin Research 31, 83-90.
Bueno, D.J., Casale, C.H., Pizzolitto, R.P., Salvano, M.A., Oliver, G., 2007. Physical adsorption of aflatoxin B1 by lactic acid bacteria and Saccharomyces cerevisiae: a theoretical model. Journal of food protection 70, 2148-2154.
Cai, M., Qian, Y., Chen, N., Ling, T., Wang, J., Jiang, H., Wang, X., Qi, K., Zhou, Y., 2020. Detoxification of aflatoxin B1 by Stenotrophomonas sp. CW117 and characterization the thermophilic degradation process. Environmental Pollution 261, 114178.
Campagnollo, F.B., Franco, L.T., Rottinghaus, G.E., Kobashigawa, E., Ledoux, D.R., Daković, A., Oliveira, C.A., 2015. In vitro evaluation of the ability of beer fermentation residue containing Saccharomyces cerevisiae to bind mycotoxins. Food research international 77, 643-648.
Cao, H., Liu, D., Mo, X., Xie, C., Yao, D., 2011. A fungal enzyme with the ability of aflatoxin B1 conversion: Purification and ESI-MS/MS identification. Microbiological Research 166, 475-483.
Caridi, A., Sidari, R., Pulvirenti, A., Blaiotta, G., 2020. Genetic improvement of wine yeasts for opposite adsorption activity of phenolics and ochratoxin A during red winemaking. Food Biotechnology 34, 352-370.
Chandra, R., 2015. Advances in biodegradation and bioremediation of industrial waste.
Chang, X., Wu, Z., Wu, S., Dai, Y., Sun, C., 2015. Degradation of ochratoxin A by Bacillus amyloliquefaciens ASAG1. Food Additives & Contaminants: Part A 32, 564-571.
Chlebicz, A., Śliżewska, K., 2020. In Vitro Detoxification of Aflatoxin B1, Deoxynivalenol, Fumonisins, T-2 Toxin and Zearalenone by Probiotic Bacteria from Genus Lactobacillus and Saccharomyces cerevisiae Yeast. Probiotics and Antimicrobial Proteins 12, 289-301.
Cho, S.M., Jeong, S.E., Lee, K.R., Sudhani, H.P., Kim, M., Hong, S.-Y., Chung, S.H., 2016. Biodegradation of ochratoxin A by Aspergillus tubingensis isolated from meju. Journal of Microbiology and Biotechnology 26, 1687-1695.
Čolović, R., Puvača, N., Cheli, F., Avantaggiato, G., Greco, D., Đuragić, O., Kos, J., Pinotti, L., 2019. Decontamination of mycotoxin-contaminated feedstuffs and compound feed. Toxins 11, 617.
Corassin, C.H., Bovo, F., Rosim, R.E., Oliveira, C.A.F.d., 2013. Efficiency of Saccharomyces cerevisiae and lactic acid bacteria strains to bind aflatoxin M1 in UHT skim milk. Food control 31, 80-83.
De Bellis, P., Tristezza, M., Haidukowski, M., Fanelli, F., Sisto, A., Mulè, G., Grieco, F., 2015. Biodegradation of ochratoxin A by bacterial strains isolated from vineyard soils. Toxins 7, 5079-5093.
De Boevre, M., Di Mavungu, J.D., Landschoot, S., Audenaert, K., Eeckhout, M., Maene, P., Haesaert, G., De Saeger, S., 2012. Natural occurrence of mycotoxins and their masked forms in food and feed products. World Mycotoxin Journal 5, 207-219.
Dhanasekaran, D., Shanmugapriya, S., Thajuddin, N., Panneerselvam, A., 2011. Aflatoxins and aflatoxicosis in human and animals. Aflatoxins- Biochemistry and Molecular Biology 10, 221-254.
Dobritzsch, D., Wang, H., Schneider, G., Yu, S., 2014. Structural and functional characterization of ochratoxinase, a novel mycotoxin-degrading enzyme. Biochemical Journal 462, 441-452.
El-Nezami, H., Mykkänen, H., Kankaanpää, P., Salminen, S., Ahokas, J., 2000. Ability of Lactobacillus and Propionibacterium strains to remove aflatoxin B1 from the chicken duodenum. Journal of food protection 63, 549-552.
El-Nezami, H., Polychronaki, N., Salminen, S., Mykkänen, H., 2002. Binding rather than metabolism may explain the interaction of two food-grade Lactobacillus strains with Zearalenone and Its Derivative ɑ́-Zearalenol. Applied and environmental microbiology 68, 3545-3549.
Fang, Q.a., Du, M., Chen, J., Liu, T., Zheng, Y., Liao, Z., Zhong, Q., Wang, L., Fang, X., Wang, J., 2020. Degradation and detoxification of aflatoxin B1 by tea-derived Aspergillus niger RAF106. Toxins 12, 777.
Fouad, M.T., El-Shenawy, M., El-Desouky, T.A., 2021. Efficiency of Selected Lactic Acid Bacteria Isolated from some Dairy Products on Aflatoxin [B.sub.1] and Ochratoxin A. Journal of Pure and Applied Microbiology 15, 312+.
Gao, F., Jiang, L.-p., Chen, M., Geng, C.-y., Yang, G., Ji, F., Zhong, L.-f., Liu, X.-f., 2013. Genotoxic effects induced by zearalenone in a human embryonic kidney cell line. Mutation Research/Genetic Toxicology and Environmental Mutagenesis 755, 6-10.
Gao, Y.-N., Wang, Z.-W., Yang, X., Wang, J.-Q., Zheng, N., 2023. Aflatoxin M1 and ochratoxin A induce a competitive endogenous RNA regulatory network of intestinal immunosuppression by whole transcriptome analysis. Science of The Total Environment 854, 158777.
Gil-Serna, J., Patiño, B., Cortés, L., González-Jaén, M.T., Vázquez, C., 2011. Mechanisms involved in reduction of ochratoxin A produced by Aspergillus westerdijkiae using Debaryomyces hansenii CYC 1244. International journal of food microbiology 151, 113-118.
Golge, O., Kabak, B., 2020. Occurrence of deoxynivalenol and zearalenone in cereals and cereal products from Turkey. Food Control 110, 106982.
Gonçalves, B.L., Rosim, R.E., de Oliveira, C.A.F., Corassin, C.H., 2015. The in vitro ability of different Saccharomyces cerevisiae–based products to bind aflatoxin B1. Food control 47, 298-300.
Gong, Y.Y., Watson, S., Routledge, M.N., 2016. Aflatoxin exposure and associated human health effects, a review of epidemiological studies. Food safety 4, 14-27.
Gratz, S., Wu, Q., El-Nezami, H., Juvonen, R., Mykkänen, H., Turner, P., 2007. Lactobacillus rhamnosus strain GG reduces aflatoxin B1 transport, metabolism, and toxicity in Caco-2 cells. Applied and environmental microbiology 73, 3958-3964.
Grenier, B., Loureiro‐Bracarense, A.P., Leslie, J.F., Oswald, I.P., 2014. Physical and chemical methods for mycotoxin decontamination in maize. Mycotoxin reduction in grain chains, 116-129.
Guan, S., Zhao, L., Ma, Q., Zhou, T., Wang, N., Hu, X., Ji, C., 2010. In vitro efficacy of Myxococcus fulvus ANSM068 to biotransform aflatoxin B1. International Journal of Molecular Sciences 11, 4063-4079.
Gupta, R.C., 2012. Veterinary toxicology: basic and clinical principles. Academic press. Haq, M., Gonzalez, N., Mintz, K., Jaja-Chimedza, A., De Jesus, C.L., Lydon, C., Welch, A.Z., Berry, J.P., 2016. Teratogenicity of ochratoxin A and the degradation product, ochratoxin α, in the zebrafish (Danio rerio) embryo model of vertebrate development. Toxins 8, 40.
Haskard, C.A., El-Nezami, H.S., Kankaanpää, P.E., Salminen, S., Ahokas, J.T., 2001. Surface binding of aflatoxin B1 by lactic acid bacteria. Applied and environmental microbiology 67, 3086-3091.
Hathout, A., Abdel-Nasser, A., 2022. The Efficiency of Saccharomyces Cerevisiae as an Antifungal and Antimycotoxigenic Agent.
Heussner, A.H., Bingle, L.E., 2015. Comparative ochratoxin toxicity: A review of the available data. Toxins 7, 4253-4282.
Hibi, D., Suzuki, Y., Ishii, Y., Jin, M., Watanabe, M., Sugita-Konishi, Y., Yanai, T., Nohmi, T., Nishikawa, A., Umemura, T., 2011. Site-Specific In V ivo Mutagenicity in the Kidney of gpt Delta Rats Given a Carcinogenic Dose of Ochratoxin A. Toxicological Sciences 122, 406-414.
Huang, L., Duan, C., Zhao, Y., Gao, L., Niu, C., Xu, J., Li, S., 2017. Reduction of aflatoxin B1 toxicity by Lactobacillus plantarum C88: a potential probiotic strain isolated from Chinese traditional fermented food “tofu”. PloS one 12, e0170109.
Jebali, R., Abbès, S., Salah-Abbès, J.B., Younes, R.B., Haous, Z., Oueslati, R., 2015. Ability of Lactobacillus plantarum MON03 to mitigate aflatoxins (B1 and M1) immunotoxicities in mice. Journal of immunotoxicology 12, 290-299.
Ju, J., Tinyiro, S.E., Yao, W., Yu, H., Guo, Y., Qian, H., Xie, Y., 2019. The ability of Bacillus subtilis and Bacillus natto to degrade zearalenone and its application in food. Journal of Food Processing and Preservation 43, e14122.
Keller, L., Abrunhosa, L., Keller, K., Rosa, C.A., Cavaglieri, L., Venâncio, A., 2015. Zearalenone and Its Derivatives α-Zearalenol and β-Zearalenol Decontamination by Saccharomyces cerevisiae Strains Isolated from Bovine Forage. Toxins 7, 3297-3308.
Kew, M.C., 2013. Aflatoxins as a cause of hepatocellular carcinoma. Journal of Gastrointestinal & Liver Diseases 22.
Khattab, A., Ibrahim, M., El-Kady, A., 2018. Ochratoxin A biosorption onto genetically improved of Lactobacillus delbrueckii mutants. International Food Research Journal 25.
Khoi, C.-S., Chen, J.-H., Lin, T.-Y., Chiang, C.-K., Hung, K.-Y., 2021. Ochratoxin A-induced nephrotoxicity: Up-to-date evidence. International Journal of Molecular Sciences 22, 11237.
Koletsi, P., Schrama, J.W., Graat, E.A., Wiegertjes, G.F., Lyons, P., Pietsch, C., 2021. The Occurrence of Mycotoxins in Raw Materials and Fish Feeds in Europe and the Potential Effects of Deoxynivalenol (DON) on the Health and Growth of Farmed Fish Species—A Review. Toxins 13, 403.
Kriszt, R., Krifaton, C., Szoboszlay, S., Cserháti, M., Kriszt, B., Kukolya, J., Czéh, Á., Fehér-Tóth, S., Török, L., Szőke, Z., 2012. A new zearalenone biodegradation strategy using non-pathogenic Rhodococcus pyridinivorans K408 strain.
Król, A., Pomastowski, P., Rafińska, K., Railean-Plugaru, V., Walczak, J., Buszewski, B., 2017. Microbiology neutralization of zearalenone using Lactococcus.
Król, A., Pomastowski, P., Rafińska, K., Railean-Plugaru, V., Walczak, J., Buszewski, B., 2018. Microbiology neutralization of zearalenone using Lactococcus lactis and Bifidobacterium sp. Analytical and bioanalytical chemistry 410, 943-952.
Kucukcakan, B., Hayrulai-Musliu, Z., 2015. Challenging role of dietary aflatoxin B1 exposure and hepatitis B infection on risk of hepatocellular carcinoma. Open access Macedonian journal of medical sciences 3, 363-369.
Lee, A., Cheng, K.-C., Liu, J.-R., 2017. Isolation and characterization of a Bacillus amyloliquefaciens strain with zearalenone removal ability and its probiotic potential. PloS one 12, e0182220.
Lei, Y., Zhao, L., Ma, Q., Zhang, J., Zhou, T., Gao, C., Ji, C., 2014. Degradation of zearalenone in swine feed and feed ingredients by Bacillus subtilis ANSB01G. World Mycotoxin Journal 7, 143-151.
Liu, Y., Mao, H., Hu, C., Tron, T., Lin, J., Wang, J., Sun, B., 2020. Molecular docking studies and in vitro degradation of four aflatoxins (AFB1, AFB2, AFG1, and AFG2) by a recombinant laccase from Saccharomyces cerevisiae. Journal of food science 85, 1353-1360.
Liuzzi, V.C., Fanelli, F., Tristezza, M., Haidukowski, M., Picardi, E., Manzari, C., Lionetti, C., Grieco, F., Logrieco, A.F., Thon, M.R., 2017. Transcriptional analysis of Acinetobacter sp. neg1 capable of degrading ochratoxin A. Frontiers in microbiology 7, 2162.
Loi, M., Fanelli, F., Cimmarusti, M.T., Mirabelli, V., Haidukowski, M., Logrieco, A.F., Caliandro, R., Mule, G., 2018. In vitro single and combined mycotoxins degradation by Ery4 laccase from Pleurotus eryngii and redox mediators. Food Control 90, 401-406.
Luz, C., Ferrer, J., Mañes, J., Meca, G., 2018. Toxicity reduction of ochratoxin A by lactic acid bacteria. Food and Chemical Toxicology 112, 60-66. Marin, D.E., Taranu, I., 2015. Ochratoxin A and its effects on immunity. Toxin Reviews 34, 11-20.
Marroquín-Cardona, A., Johnson, N., Phillips, T., Hayes, A., 2014. Mycotoxins in a changing global environment–a review. Food and Chemical Toxicology 69, 220-230.
Mohsenzadeh, M.S., Hedayati, N., Riahi-Zanjani, B., Karimi, G., 2016. Immunosuppression following dietary aflatoxin B1 exposure: a review of the existing evidence. Toxin Reviews 35, 121-127.
Moll, D., 2019. Enzyme technology for detoxification of mycotoxins in animal feed. Industrial Enzyme Applications, 219-254.
Nazareth, T.d.M., Luz, C., Torrijos, R., Quiles, J.M., Luciano, F.B., Mañes, J., Meca, G., 2019. Potential application of lactic acid bacteria to reduce aflatoxin B1 and fumonisin B1 occurrence on corn kernels and corn ears. Toxins 12, 21.
Niderkorn, V., Morgavi, D.P., Pujos, E., Tissandier, A., Boudra, H., 2007. Screening of fermentative bacteria for their ability to bind and biotransform deoxynivalenol, zearalenone and fumonisins in an in vitro simulated corn silage model. Food Additives & Contaminants 24, 406-415.
Ogunade, I., Martinez-Tuppia, C., Queiroz, O., Jiang, Y., Drouin, P., Wu, F., Vyas, D., Adesogan, A., 2018. Silage review: Mycotoxins in silage: Occurrence, effects, prevention, and mitigation. Journal of dairy science 101, 4034-4059.
Pankaj, S., Shi, H., Keener, K.M., 2018. A review of novel physical and chemical decontamination technologies for aflatoxin in food. Trends in Food Science & Technology 71, 73-83.
Patriarca, A., Pinto, V.F., 2017. Prevalence of mycotoxins in foods and decontamination. Current Opinion in Food Science 14, 50-60.
Peltonen, K., El-Nezami, H., Haskard, C., Ahokas, J., Salminen, S., 2001. Aflatoxin B1 binding by dairy strains of lactic acid bacteria and bifidobacteria. Journal of dairy science 84, 2152-2156.
Petruzzi, L., Bevilacqua, A., Baiano, A., Beneduce, L., Corbo, M.R., Sinigaglia, M., 2014. In vitro removal of ochratoxin A by two strains of Saccharomyces cerevisiae and their performances under fermentative and stressing conditions. Journal of Applied Microbiology 116, 60-70.
Pfohl-Leszkowicz, A., Manderville, R.A., 2012. An update on direct genotoxicity as a molecular mechanism of ochratoxin a carcinogenicity. Chemical research in toxicology 25, 252-262.
Piotrowska, M., 2014. The adsorption of ochratoxin A by Lactobacillus species. Toxins 6, 2826-2839.
Piotrowska, M., Nowak, A., Czyzowska, A., 2013. Removal of ochratoxin A by wine Saccharomyces cerevisiae strains. European Food Research and Technology 236, 441-447.
Raju, M., Devegowda, G., 2000. Influence of esterified-glucomannan on performance and organ morphology, serum biochemistry and haematology in broilers exposed to individual and combined mycotoxicosis (aflatoxin, ochratoxin and T-2 toxin). British poultry science 41, 640-650.
Rao, K.R., Vipin, A., Hariprasad, P., Appaiah, K.A., Venkateswaran, G., 2017. Biological detoxification of Aflatoxin B1 by Bacillus licheniformis CFR1. Food Control 71, 234-241.
Rieswijk, L., Claessen, S.M., Bekers, O., van Herwijnen, M., Theunissen, D.H., Jennen, D.G., de Kok, T.M., Kleinjans, J.C., van Breda, S.G., 2016. Aflatoxin B1 induces persistent epigenomic effects in primary human hepatocytes associated with hepatocellular carcinoma. Toxicology 350, 31-39.
Rogowska, A., Pomastowski, P., Walczak, J., Railean-Plugaru, V., Rudnicka, J., Buszewski, B., 2019. Investigation of zearalenone adsorption and biotransformation by microorganisms cultured under cellular stress conditions. Toxins 11, 463.
Rotimi, O.A., Rotimi, S.O., Duru, C.U., Ebebeinwe, O.J., Abiodun, A.O., Oyeniyi, B.O., Faduyile, F.A., 2017. Acute aflatoxin B1–Induced hepatotoxicity alters gene expression and disrupts lipid and lipoprotein metabolism in rats. Toxicology reports 4, 408-414.
Sadiq, F.A., Yan, B., Tian, F., Zhao, J., Zhang, H., Chen, W., 2019. Lactic acid bacteria as antifungal and anti‐mycotoxigenic agents: a comprehensive review. Comprehensive Reviews in Food Science and Food Safety 18, 1403-1436.
Sandlin, N., Russell Kish, D., Kim, J., Zaccaria, M., Momeni, B., 2022. Current and emerging tools of computational biology to improve the detoxification of mycotoxins. Applied and Environmental Microbiology 88, e02102-02121.
Sangare, L., Zhao, Y., Folly, Y.M.E., Chang, J., Li, J., Selvaraj, J.N., Xing, F., Zhou, L., Wang, Y., Liu, Y., 2014. Aflatoxin B1 degradation by a Pseudomonas strain. Toxins 6, 3028-3040.
Sangsila, A., Faucet-Marquis, V., Pfohl-Leszkowicz, A., Itsaranuwat, P., 2016. Detoxification of zearalenone by Lactobacillus pentosus strains. Food Control 62, 187-192.
Scarpari, M., Bello, C., Pietricola, C., Zaccaria, M., Bertocchi, L., Angelucci, A., Ricciardi, M.R., Scala, V., Parroni, A., Fabbri, A.A., 2014. Aflatoxin control in maize by Trametes versicolor. Toxins 6, 3426-3437.
Schwartz, P., Bucheli, T.D., Wettstein, F.E., Burkhardt‐Holm, P., 2013. Life‐cycle exposure to the estrogenic mycotoxin zearalenone affects zebrafish (Danio rerio) development and reproduction. Environmental Toxicology 28, 276-289.
Shang, L., Bai, X., Chen, C., Liu, L., Li, M., Xia, X., Wang, Y., 2019. Isolation and identification of a Bacillus megaterium strain with ochratoxin A removal ability and antifungal activity. Food Control 106, 106743.
Shang, Q., Yang, Z., Yang, W., Li, Z., Zhang, G., Jiang, S., 2016. Toxicity of mycotoxins from contaminated corn with or withoutyeast cell wall adsorbent on broiler chickens. Asian-Australasian Journal of Animal Sciences 29, 674.
Shetty, P.H., Jespersen, L., 2006. Saccharomyces cerevisiae and lactic acid bacteria as potential mycotoxin decontaminating agents. Trends in food science & technology 17, 48-55.
Shi, L., Liang, Z., Li, J., Hao, J., Xu, Y., Huang, K., Tian, J., He, X., Xu, W., 2014. Ochratoxin A biocontrol and biodegradation by Bacillus subtilis CW 14. Journal of the Science of Food and Agriculture 94, 1879-1885.
Shi, L., Liang, Z., Xu, S., Zheng, H., Huang, K., 2013. Adsorption and degradation of ochratoxin A by Bacillus licheniformis Sl-1. Journal of Agricultural Biotechnology 21, 1420-1425.
Shu, X., Wang, Y., Zhou, Q., Li, M., Hu, H., Ma, Y., Chen, X., Ni, J., Zhao, W., Huang, S., 2018. Biological degradation of aflatoxin B1 by cell-free extracts of Bacillus velezensis DY3108 with broad pH stability and excellent thermostability. Toxins 10, 330.
Shukla, S., Park, J.H., Kim, M., 2020. Efficient, safe, renewable, and industrially feasible strategy employing Bacillus subtilis with alginate bead composite for the reduction of ochratoxin A from wine. Journal of Cleaner Production 242, 118344.
Smith, M.-C., Madec, S., Coton, E., Hymery, N., 2016. Natural co-occurrence of mycotoxins in foods and feeds and their in vitro combined toxicological effects. Toxins 8, 94.
Song, J., Zhang, S., Xie, Y., Li, Q., 2019. Purification and characteristics of an aflatoxin B1 degradation enzyme isolated from Pseudomonas aeruginosa. FEMS microbiology letters 366, fnz034.
Song, Y., Wang, Y., Guo, Y., Qiao, Y., Ma, Q., Ji, C., Zhao, L., 2021. Degradation of zearalenone and aflatoxin B1 by Lac2 from Pleurotus pulmonarius in the presence of mediators. Toxicon 201, 1-8.
Streit, E., Schatzmayr, G., Tassis, P., Tzika, E., Marin, D., Taranu, I., Tabuc, C., Nicolau, A., Aprodu, I., Puel, O., 2012. Current situation of mycotoxin contamination and co-occurrence in animal feed—Focus on Europe. Toxins 4, 788-809.
Taheur, F.B., Fedhila, K., Chaieb, K., Kouidhi, B., Bakhrouf, A., Abrunhosa, L., 2017. Adsorption of aflatoxin B1, zearalenone and ochratoxin A by microorganisms isolated from Kefir grains. International Journal of Food Microbiology 251, 1-7.
Takahashi-Ando, N., Kimura, M., Kakeya, H., Osada, H., Yamaguchi, I., 2002. A novel lactonohydrolase responsible for the detoxification of zearalenone: enzyme purification and gene cloning. Biochemical journal 365, 1-6.
Taroub, B., Salma, L., Manel, Z., Ouzari, H.-I., Hamdi, Z., Moktar, H., 2019. Isolation of lactic acid bacteria from grape fruit: antifungal activities, probiotic properties, and in vitro detoxification of ochratoxin A. Annals of Microbiology 69, 17-27.
Taylor, M.C., Jackson, C.J., Tattersall, D.B., French, N., Peat, T.S., Newman, J., Briggs, L.J., Lapalikar, G.V., Campbell, P.M., Scott, C., 2010. Identification and characterization of two families of F420H2 dependent reductases from Mycobacteria that catalyse aflatoxin degradation. Molecular microbiology 78, 561-575.
Tola, M., Kebede, B., 2016. Occurrence, importance and control of mycotoxins: A review. Cogent Food & Agriculture 2, 1191103. Utermark, J., Karlovsky, P., 2007. Role of zearalenone lactonase in protection of Gliocladium roseum from fungitoxic effects of the mycotoxin zearalenone. Applied and environmental microbiology 73, 637-642.
Varga, J., Frisvad, J.C., Samson, R., 2011. Two new aflatoxin producing species, and an overview of Aspergillus section Flavi. Studies in mycology 69, 57-80.
Vega, M.F., Dieguez, S.N., Riccio, B., Aranguren, S., Giordano, A., Denzoin, L., Soraci, A.L., Tapia, M.O., Ross, R., Apás, A., 2017. Zearalenone adsorption capacity of lactic acid bacteria isolated from pigs. brazilian journal of microbiology 48, 715-723.
Vila-Donat, P., Marín, S., Sanchis, V., Ramos, A., 2018. A review of the mycotoxin adsorbing agents, with an emphasis on their multi-binding capacity, for animal feed decontamination. Food and chemical toxicology 114, 246-259.
Wall-Martínez, H.A., Pascari, X., Bigordà, A., Ramos, A.J., Marín, S., Sanchis, V., 2019. The fate of Fusarium mycotoxins (deoxynivalenol and zearalenone) through wort fermenting by Saccharomyces yeasts (S. cerevisiae and S. pastorianus). Food Research International 126, 108587.
Wang, J., Ogata, M., Hirai, H., Kawagishi, H., 2011. Detoxification of aflatoxin B1 by manganese peroxidase from the white-rot fungus Phanerochaete sordida YK-624. FEMS microbiology letters 314, 164-169.
Wang, J., Xie, Y., 2020. Review on microbial degradation of zearalenone and aflatoxins. Grain & Oil Science and Technology 3, 117-125.
Wang, L., Wu, J., Liu, Z., Shi, Y., Liu, J., Xu, X., Hao, S., Mu, P., Deng, F., Deng, Y., 2019a. Aflatoxin B1 degradation and detoxification by Escherichia coli CG1061 isolated from chicken cecum. Frontiers in pharmacology 9, 1548.
Wang, N., Wu, W., Pan, J., Long, M., 2019b. Detoxification strategies for zearalenone using microorganisms: A review. Microorganisms 7, 208.
Wang, X., Bai, Y., Huang, H., Tu, T., Wang, Y., Wang, Y., Luo, H., Yao, B., Su, X., 2019c. Degradation of aflatoxin B1 and zearalenone by bacterial and fungal laccases in presence of structurally defined chemicals and complex natural mediators. Toxins 11, 609.
Wang, X., Qin, X., Hao, Z., Luo, H., Yao, B., Su, X., 2019d. Degradation of four major mycotoxins by eight manganese peroxidases in presence of a dicarboxylic acid. Toxins 11, 566.
Woźny, M., Dobosz, S., Hliwa, P., Gomułka, P., Król, J., Obremski, K., Blahova, J., Svobodova, Z., Michalik, O., Ocalewicz, K., 2020. Feed-borne exposure to zearalenone impairs reproduction of rainbow trout. Aquaculture 528, 735522.
Wu, N., Ou, W., Zhang, Z., Wang, Y., Xu, Q., Huang, H., 2021. Recent advances in detoxification strategies for zearalenone contamination in food and feed. Chinese Journal of Chemical Engineering 30, 168-177.
Wu, Y.-Z., Lu, F.-P., Jiang, H.-L., Tan, C.-P., Yao, D.-S., Xie, C.-F., Liu, D.-L., 2015. The furofuran-ring selectivity, hydrogen peroxide-production and low Km value are the three elements for highly effective detoxification of aflatoxin oxidase. Food and Chemical Toxicology 76, 125-131.
Xiong, D., Wen, J., Lu, G., Li, T., Long, M., 2022. Isolation, purification, and characterization of a laccase degrading aflatoxin b1 from Bacillus amyloliquefaciens B10. Toxins 14, 250.
Xiong, K., Wang, X.l., Zhi, H.W., Sun, B.G., Li, X.T., 2017. Identification and safety evaluation of a product from the biodegradation of ochratoxin A by an Aspergillus strain. Journal of the Science of Food and Agriculture 97, 434-443.
Xiong, K., Zhi, H.-w., Liu, J.-y., Wang, X.-y., Zhao, Z.-y., Pei, P.-g., Deng, L., Xiong, S.-y., 2021. Detoxification of Ochratoxin A by a novel Aspergillus oryzae strain and optimization of its biodegradation. Revista argentina de microbiología 53, 48-58.
Xu, H., Wang, L., Sun, J., Wang, L., Guo, H., Ye, Y., Sun, X., 2022. Microbial detoxification of mycotoxins in food and feed. Critical reviews in food science and nutrition 62, 4951-4969.
Xu, J., Wang, H., Zhu, Z., Ji, F., Yin, X., Hong, Q., Shi, J., 2016. Isolation and characterization of Bacillus amyloliquefaciens ZDS-1: Exploring the degradation of Zearalenone by Bacillus spp. Food Control 68, 244-250.
Xu, X., Pang, M., Liu, J., Wang, Y., Wu, X., Huang, K., Liang, Z., 2021. Genome mining reveals the genes of carboxypeptidase for OTA-detoxification in Bacillus subtilis CW14. International journal of biological macromolecules 186, 800-810.
Yi, P.-J., Pai, C.-K., Liu, J.-R., 2011. Isolation and characterization of a Bacillus licheniformis strain capable of degrading zearalenone. World Journal of Microbiology and Biotechnology 27, 1035-1043.
Yiannikouris, A., FRANCois, J., Poughon, L., Dussap, C.-G., Bertin, G., Jeminet, G., Jouany, J.-P., 2004. Alkali extraction of β-D-glucans from Saccharomyces cerevisiae cell wall and study of their adsorptive properties toward zearalenone. Journal of Agricultural and Food Chemistry 52, 3666-3673.
Yu, Y., Qiu, L., Wu, H., Tang, Y., Lai, F., Yu, Y., 2011a. Oxidation of zearalenone by extracellular enzymes from Acinetobacter sp. SM04 into smaller estrogenic products. World Journal of Microbiology and Biotechnology 27, 2675-2681.
Yu, Y., Qiu, L., Wu, H., Tang, Y., Yu, Y., Li, X., Liu, D., 2011b. Degradation of zearalenone by the extracellular extracts of Acinetobacter sp. SM04 liquid cultures. Biodegradation 22, 613-622.
Zhang, H., Dong, M., Yang, Q., Apaliya, M.T., Li, J., Zhang, X., 2016. Biodegradation of zearalenone by Saccharomyces cerevisiae: Possible involvement of ZEN responsive proteins of the yeast. Journal of Proteomics 143, 416-423.
Zhang, Y., Li, Z., Lu, Y., Zhang, J., Sun, Y., Zhou, J., Tu, T., Gong, W., Sun, W., Wang, Y., 2022. Characterization of Bacillus velezensis E2 with abilities to degrade ochratoxin A and biocontrol against Aspergillus westerdijkiae fc-1. Toxicon 216, 125-131.
Zhang, Y., Wu, D., Su, Y., Xie, B., 2021. Occurrence, influence and removal strategies of mycotoxins, antibiotics and microplastics in anaerobic digestion treating food waste and co-digestive biosolids: a critical review. Bioresource Technology 330, 124987.
Zhao, M., Wang, X., Xu, S., Yuan, G., Shi, X., Liang, Z., 2020. Degradation of ochratoxin A by supernatant and ochratoxinase of Aspergillus niger W-35 isolated from cereals. World Mycotoxin Journal 13, 287-298.




 Micotoxicosis prevention
Micotoxicosis prevention